BIOL 570. Lab 6. Binomial distribution and the binomial test
Review the quick summary of the chapter on Canvas, and you may want to look at the steps in hypothesis testing handout on Canvas.
Nearly all biological populations are genetically variable. Today we will focus on a genetically variable morphological trait that is easy to score: whether a person’s ear lobe is “free” or “attached” (see below). The typical frequency of the “free” phenotype is 0.65, although there is variation around the world.
Step 1: Just to get a quick-to-obtain data set, your TA will take data on the number of people with free and attached ear lobes in your class and record this information on the board.
Step 2:
Recall that the probability of obtaining
X
successes and is defined as:
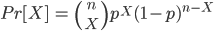 where
where
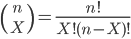 .
.
We will arbitrarily define a “success” as a person with a “free” ear lobe. Imagine you want to know: Given that the probability of a “free” ear lobe is 0.65, what is the probability of collecting your sample (i.e. X people with a “free” ear lobe in a group of n = _______ people?)
Question 1. What is this probability? To answer this question, state the numerical value of n, p, and X, write out the appropriate binomial formula for this problem, and then solve the formula.
Now [a]
check your calculations for Question 1 using RStudio.
You can do this by setting
x,
n, and
p
variables to correspond to the values that you are interested in.